Small (or short) interfering RNA (siRNA) is the most commonly used RNA interference (RNAi) tool for inducing short-term silencing of protein coding genes.
What is siRNA?
siRNA is a synthetic RNA duplex designed to specifically target a particular mRNA for degradation. While siRNA provides the opportunity to induce gene knockdown in a variety of cell lines, their utility is limited to cells that are amenable to transfection of synthetic oligonucleotides.
Since siRNAs achieve transient silencing, experiments are limited to relatively short time frames on the order of 2-4 days. siRNAs can also be used for knockdown of non-protein coding genes, such as long noncoding RNAs (lncRNA).
How does siRNA work?
siRNAs consist of two RNA strands, an antisense (or guide) strand and a sense (or passenger) strand, which form a duplex 19 to 25 bp in length with 3' dinucleotide overhangs (Figure 1).
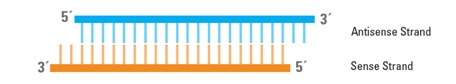
Structure of siRNA
Synthetic siRNAs are most commonly generated through solid-phase chemical synthesis methods (such as patented 2'-ACE chemistry) which provide highly pure, stable, and
readily modified siRNAs. Small double-strand siRNAs are transfected into cells where the guide strand is loaded into RISC. This activated protein and nucleic acid complex can then elicit gene silencing by binding, through perfect complementarity, to a single target mRNA sequence, thereby targeting it for cleavage and degradation.
How is siRNA delivered to a cell?
siRNA must be transfected into cells either by cationic lipid or polymer-based transfection reagents, electroporation (physical delivery via plasma membrane holes created by an electrical field), or adding chemical modifications to the duplex to aid in uptake by the cell.
| Delivery Mode | Advantages | Disadvantages | |
|---|---|---|---|
| Transfection |
|
|
|
| Electroporation |
|
|
|
| Viral-mediated Delivery |
|
|
|
| Modified siRNA |
|
|
|
siRNA function
The success of RNAi experiments depends on the efficiency of gene knockdown. Early work on siRNA design established conventional guidelines for siRNA structural attributes that led to reasonable functional knockdown in specific cases1.
The properties of potent siRNAs were further refined by performing large-scale functional studies that defined thermodynamic and sequence-based rules for rational siRNA design2. These design algorithms greatly improved the reliability of identifying potent siRNA sequences. The Dharmacon SMARTselection algorithm was the first comprehensive rational design strategy applied to commercial collections. While research is continually striving to identify molecules with greater activity and specificity, siRNAs designed by SMARTselection strategies, such as siGENOME and ON-TARGETplus reagents, remain the mo mst effective reagents on thearket.
Specificity
Although the sequence complementarity-based mechanism underlying RNAi allows for target-specific gene knockdown, the same mechanism can result in unintended knockdown of genes not being directly targeted. Several strategies have been developed to mitigate these so-called "off-target" effects and ensure on-target activity.
Chemical modifications to the siRNA have been used successfully to promote preferential loading of the intended antisense (guide) strand into the RISC complex3,4 and reduce sense (passenger) strand loading and activity5,6. Further, to reduce the risk of the siRNA guide strand seed region from causing off-target effects, design algorithms can incorporate filters that exclude high-frequency seed sequences from known mammalian microRNAs7. Chemical modifications or thermodynamic-based design considerations can also be applied to the siRNA seed region to discourage undesired interactions5,8,9. Finally, the strategy of pooling several independent siRNAs that target an individual gene has been shown to reduce the total number of non-specific gene targets and the frequency of off-target phenotypes while preserving potent target gene knockdown10. All of these strategies, when combined, work efficiently to reduce off-targeting and to achieve potent and specific silencing for a successful RNAi experiment.
Applications
siRNAs are widely used to assess the individual contributions of genes to an assortment of cellular phenotypes including cytokinesis11, apoptosis12, insulin signaling13,14 and cell differentiation15.
siRNA screens have been used to identify novel pathways16 and have had significant impact in validating targets for a number of cellular processes and diseases including cancer17,18, HIV infection19 and hepatitis20. Finally, in vivo RNAi has been used for target validation studies in animal disease models and has the potential to be used for therapeutic purposes where disease-causing genes are selectively targeted and suppressed21.
Controls for siRNA experiments
Controls are an essential part of every siRNA experiment. At least three types of controls should be used in each siRNA (and RNAi) experiment: positive control, negative control and untreated control.
A well-characterized positive control allows the researcher to ensure the delivery method is sufficient to achieve effective silencing. Negative controls help to separate sequence-specific effects from the effects of experimental conditions on cellular responses. An untreated control establishes a useful baseline reference for cell phenotypes and gene expression levels.
Effective controls for RNAi - Tech Note
- Chemically synthesized siRNA reagents that target every gene in human, mouse and rat genome are available for convenient delivery in vitro.
C911 controls for siRNA screening
- For large scale siRNA screens, C911 controls can be a useful approach when carrying out in-depth hit confirmation.
References
- Elbashir, S.M., et al., Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 2001. 411: p. 494-8.
- Reynolds, A., et al., Rational siRNA design for RNA interference. Nature Biotechnology, 2004. 22(3): p. 326-330.
- Khvorova, A., A. Reynolds, and S. Jayasena, Functional siRNAs and miRNAs exhibit strand bias. Cell, 2003. 115: p. 209-216.
- Schwarz, D.S., et al., Asymmetry in the assembly of the RNAi enzyme complex. Cell, 2003. 115(2): p. 199-208.
- Jackson, A.L., et al., Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing. Rna, 2006.
- Vaish, N., et al., Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res., 2011. 39(5): p. 1823-1832.
- Birmingham, A., et al., A protocol for designing siRNAs with high functionality and specificity. Nat Protoc, 2007. 2: p. 2068-78.
- Bramsen, J.B., et al., A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res., 2010. 38(17): p. 5761-5773.
- Ui-Tei, K., et al., Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res., 2008. 36(22): p. 7100-7109.
- Kittler, R., et al., Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods, 2007. 4: p. 337-44.
- Zhou, T., et al., A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell, 2003. 5(1): p. 127-38.
- Zender, L., et al., Caspase 8 small interfering RNA prevents acute liver failure in mice. PNAS, 2003. 100(13): p. 7797-7802.
- Hsieh, A.C., et al., A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: determinants of gene silencing for use in cell-based screens. Nucleic Acids Res, 2004. 32: p. 893-901.
- Jiang, Z.Y., et al., Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. PNAS, 2003. 100: p. 7569-7574.
- Kurisaki, K., et al., Nuclear Factor YY1 Inhibits Transforming Growth Factor {beta}- and Bone Morphogenetic Protein-Induced Cell Differentiation. Mol. Cell. Biol., 2003. 23: p. 4494-4510.
- Struwe, W.B. and C.E. Warren, High-throughput RNAi screening for N-glycosylation dependent loci in Caenorhabditis elegans. Methods Enzymol, 2010. 480: p. 477-93.
- Whitehurst, A.W., et al., Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature, 2007. 446(7137): p. 815-9.
- Bric, A., et al., Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell, 2009. 16(4): p. 324-35.
- Kok, K.H., T. Lei, and D.Y. Jin, siRNA and shRNA screens advance key understanding of host factors required for HIV-1 replication. Retrovirology, 2009. 6: p. 78.
- Li, Q., et al., A genome-wide genetic screen for host factors required for hepatitis C virus propagation. PNAS, 2009. 106: p. 16410-16415.
- Dominska, M. and D.M. Dykxhoorn, Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci, 2010. 123(Pt 8): p. 1183-9.
Which siRNA tool is right for your needs?
siRNA selection guide
We offer a wide selection of predesigned siRNA product lines and formats. This quick selection guide will help to determine the best option for your particular needs
Order Products
ON-TARGETplus siRNA
Guaranteed gene silencing and patented modifications to reduce off-targets – pre-designed ON-TARGETplus siRNA are available as individual or in SMARTpool format for human, mouse or rat genes
Accell siRNA
A novel siRNA for difficult-to-transfect cells, modified to require no transfection reagent or viral vector for delivery
siGENOME siRNA
Expert siRNA design and guaranteed gene silencing provide a value-priced option for high quality RNAi results
Lincode siRNA
Expert siRNA design and specificity-enhancing modifications for effective knockdown of human long non-coding RNA
Cherry-pick library design tool
Design and order your own custom siRNA mini-library to screen you genes of interest
Custom siRNA synthesis
Lacinia quis vel eros donec ac odio tempor orci.
Dharmacon Transfection Products
Reagents optimized for your specific application and cell line
Helpful Resources
Accell siRNA reagents: achieving long-term gene silencing - Application note
Developing an understanding of a gene’s contribution to a particular phenotype is problematic when the protein has an extended half life
Basic siRNA resuspension - Protocol
This protocol outlines resuspension of siRNA and includes answers to frequently asked questions
Off-target effects: disturbing the silence of RNA interference - Application note
Bioinformatics, novel chemical modifications, and siRNA pooling significantly decrease off-target effects
