Genome engineering with homology-directed repair
Homologous recombination is the exchange of DNA sequence information through the use of sequence homology. Homology-directed repair (HDR) is a process of homologous recombination where a DNA template is used to provide the homology necessary for precise repair of a double-strand break (DSB). This template can come from within the cell during the late stage of S phase and from the G2 phase of the cell cycle, when sister chromatids are available prior to the completion of mitosis. Additionally, exogenous repair templates can be delivered into a cell, most often in the form of a synthetic, single-strand DNA donor oligo or DNA donor plasmid, to generate a precise change in the genome.
Use of single-strand DNA donor oligos for HDR
Single-strand DNA donor oligos are delivered into a cell to insert or change short sequences (SNPs, amino acid substitutions, epitope tags, etc.) of DNA in the endogenous genomic target region. The benefits of using a synthetic DNA donor oligo is that no cloning is required to generate the donor template and DNA modifications can be added during synthesis for different applications, such as increased resistance to nucleases. Traditionally, the maximum insert length recommended for use with a DNA donor oligo is ~ 50 nucleotides.
Design considerations for DNA donor oligos for HDR
To design a DNA donor oligo for precise gene editing using HDR, one must consider the distance of the intended HDR modification site (or insertion site) from the Cas9 cut site, the crRNA for a specific insertion site, the type of HDR modification, the length of homology arms, and disruption of the CRISPR targeting site if it is present in the DNA donor oligo. Here we describe designing DNA donor oligos for use with the CRISPR-Cas9 system; general principles can be applied to any site-directed nuclease.
Insertion site and cut site distance
In mammalian cells, HDR has highest efficiency when the insertion site and the DSB are within ten nucleotides from one another, and HDR efficiencies drop quickly as this distance increases1.
Identifying candidate crRNAs
The DNA region 20 nt upstream and downstream from the desired insertion site should be assessed for PAM sites on both strands, and 3-5 candidate crRNAs should be tested for cutting efficiency with a DNA mismatch detection assay.
Selecting crRNAs for maximal HDR efficiency at a specific insertion site
Since HDR efficiency is relatively low compared to non-homologous end joining (NHEJ) repair of DSBs, it is important to choose a crRNA with the highest possible DNA cutting efficiency. An efficiency of at least 25% is recommended; the higher the number of DSBs that Cas9 will generate, the more cut sites available to the HDR machinery for repair with the DNA donor oligo.
Not every insertion site will have PAM sequences within the surrounding 20 nt, or the assessed crRNA efficiency with available PAM sites might be very low. In these circumstances, choose the most active crRNA for your given gene target region that is as close as possible to the intended insertion site for the best chance of detecting an HDR gene modification at your genomic cut site. Again be aware that HDR has highest efficiency when the insertion site and the DSB are within ten nucleotides from one another.
Retrieving a genomic sequence to design a synthetic DNA donor oligo
An annotated genomic sequence is required to begin designing a DNA donor oligo. When selecting the genomic sequence, it is essential to retrieve a genomic sequence (introns and exons) so that your DNA template correctly repairs the intended HDR modification site.
Length of homology arms for a synthetic DNA donor oligo
When designing a DNA donor oligo to replace a nucleotide or to insert an epitope tag2, homology arms need to flank the desired HDR modification and be specific to the site where the HDR modification should take place (Figure 1). The length of each homology arm of the donor oligo can vary from short (30-40 nt) to long (70-90 nt) and typically are symmetric in length around the desired insert3,4. We recommend using 30-40 nucleotides for both the 5' and 3' homology arms when using single-strand DNA donor oligos5.It has been demonstrated3,5 that the addition of two phosphorothioates on each end of the DNA oligo increases HDR efficiency over non-phosphorothioated DNA donor oligos.
Designing DNA donor oligos for HDR
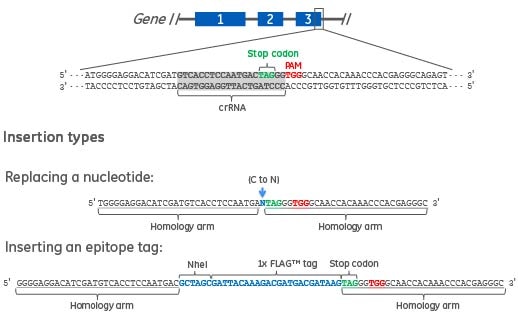
Figure 1. Two types of DNA donor oligos are shown. The first is a 61 nucleotide DNA donor oligo to replace a single nucleotide in the target gene. The homology arms specific to the gene target (underlined) are flanking the nucleotide to be changed (blue text). The second is a 90 nucleotide DNA donor oligo to create a C-terminal epitope tag (e.g. FLAG) and a RFLP detection site (e.g. NheI). The homology arms specific to the gene target (underlined) are flanking the epitope tag and NheI restriction site (blue text).
Avoiding CRISPR-Cas9 cutting of an HDR gene locus
When designing a donor template, it is essential to disrupt the CRISPR recognition and cut site (~ 20 nucleotide target sequence and PAM), of your intended CRISPR target site when it occurs within your donor template. Failure to disrupt the CRISPR site will allow Cas9 to cut your HDR-edited gene locus after the repair has taken place. This can be accomplished with one or more of the following changes to the donor template (Figure 2): (1) design the donor oligo so that your intended DNA modification splits the 20 nucleotide targeting sequence, or a portion of the targeting sequence, and the corresponding PAM. The two homology arms of the donor oligo will surround the intended DNA modification, or (2) create nucleotide changes in the PAM or targeting region to prevent Cas9-targeted cleavage at the HDR-edited site. Either design option (1) or (2), not both, will be required to avoid Cas9 cleavage of an HDR gene locus.
Donor oligo design strategies to avoid CRISPR-Cas9 cutting after HDR-mediated editing
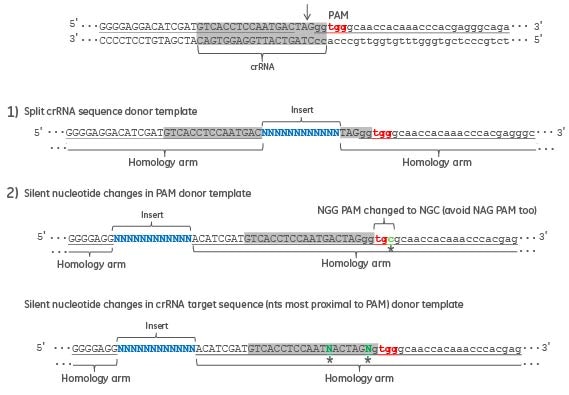
Figure 2. An example genomic sequence, with the exon sequence in uppercase and intron sequence in lower case, is displayed with the crRNA target sequence in gray. Donor oligo design strategies are presented that will avoid additional rounds of CRISPR-Cas9 cleavage and NHEJ repair after insertion of desired sequence by HDR.
Detecting HDR with restriction enzymes
It is suggested to include a restriction enzyme site, either by insertion of the restriction enzyme recognition sequence or make nucleotide changes near the insert to create a restriction enzyme site; this facilitates detection of successful HDR by PCR-based detection assays, such as restriction fragment length polymorphism (RFLP); cleavage of the PCR product at the newly introduced restriction enzyme site.
References
- B. Elliott, C. Richardson, et al., Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell Biol. 18(1), 93-101 (1998)
- Bill Brizzard., Epitope tagging Biotechniques. 44(5), 693-5 (2008)
- JB. Renaud, C. Boix, et al., Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. 14(9), 2263-72 (2016)
- FA. Ran, PD. Hsu, et al., Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity.Cell. 154(6), 1380-9 (2013)
- J. Schiel, E. Chou, et al., Homology-directed repair with Dharmacon™ Edit-R™ CRISPR-Cas9 reagents and single-strand DNA oligos.Dharmacon 2015
Order Products
Custom DNA Synthesis
Use our convenient online tools to design and order the custom DNA oligo that fits your experimental design.
CRISPR-Cas9 Gene Editing
Optimized tools for high-confidence genome engineering
CRISPR Design Tool
Design custom crRNA and sgRNA for gene editing applications
HDR Donor Designer Workflow
Design and order a custom donor oligo or plasmid for HDR-mediated gene editing in over 30 different species
Helpful Resources
Read protocol
Edit-R Cas9 mRNA, crRNA:tracrRNA and HDR donor template transfection
Read app note
Homology-directed repair with Dharmacon Edit-R CRISPR-Cas9 reagents and single-strand DNA oligos
View poster
Homology-directed repair using synthetic crRNA and tracrRNA with single-strand DNA oligos
Read app note
A CRISPR-Cas9 gene engineering workflow: generating functional knockouts using Edit-R Cas9 and synthetic crRNA and tracrRNA
Read app note
Microinjection of zebrafish embryos using Dharmacon Edit-R Cas9 Nuclease mRNA, synthetic crRNA, and tracrRNA for genome engineering
