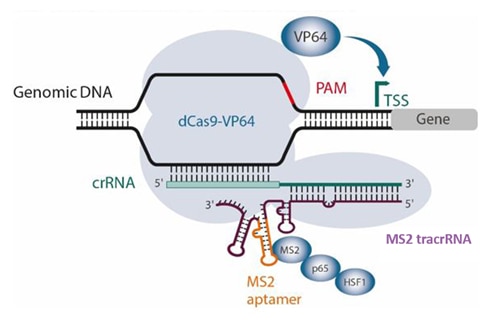
CRISPR activation (CRISPRa) repurposes the S. pyogenes CRISPR-Cas9 system. Review our CRISPRa applications page to get an overview of the technology.
In the synergistic activation mediator (SAM)1 activation system, fusion proteins dCas9-VP64 and MS2-p65-HSF1 are used in combination with an MS2 aptamer-modified guide RNA to facilitate binding of the MS2-p65-HSF1 activating module. Published expressed versions of this SAM guide RNA contain two MS2 aptamers. Synthetic versions of SAM guide RNA are challenging to synthesize, because an analogous SAM sgRNA or MS2 tracrRNA would be 158 and 141 nucleotides respectively.
Taking advantage of Horizon Discovery’s long RNA synthesis and optimization capabilities, we have developed a synthetic MS2 tracrRNA that contains a single MS2 aptamer sequence on stem loop 2 (Figure 1). When used with gene-specific CRISPRa crRNA, MS2 tracrRNA abundantly recruits the MS2-p65-HSF1 fusion to the transcriptional start site (TSS) to initiate activation. CRISPRa guide RNA designs come from a published CRISPRa v2 algorithm based on machine learning2 and enable robust over-expression of your gene(s) of interest.
Achieve robust gene activation with synthetic MS2 tracrRNA
We have assessed the activation of synthetic crRNA and MS2 tracrRNA by measuring the transcriptional fold activation for six genes of interest (Figure 2). U2OS and A375 cells stably expressing the SAM activation system (dCas9-VP64 and MS2-p65-HSF1) were transfected with synthetic MS2 tracrRNA and CRISPRa pooled crRNA using DharmaFECT transfection reagents. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. Robust activation ranging from 17 to almost 10,000-fold was observed for all six genes in both cell lines.
CRISPRa in d-Cas9-SAM expressing cells with synthetic crRNA with MS2 tracrRNA
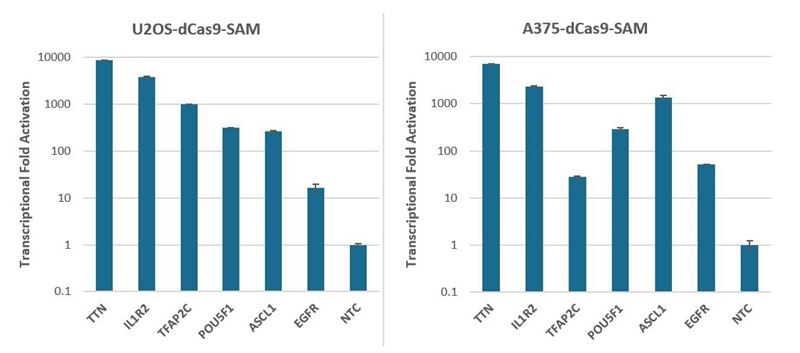
Figure 2. U2OS and A375 cells stably expressing dCas9-VP64 and MS2-p65-HSF1 were plated at 10,000 cells/well and transfected with CRISPRa pooled crRNA complexed with MS2 tracrRNA (25nM), using either DharmaFECT 4 (U2OS-dCas9-SAM) or DharmaFECT 1 (A375-dCas9-SAM) Transfection Reagent. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using PPIB as the housekeeping gene and normalized to a non-targeting control.
Synthetic crRNA with MS2 tracrRNA offers improved activation versus expressed SAM sgRNA
All publications to date for SAM-mediated activation have utilized expressed SAM sgRNA. We investigated how levels of activation compare when using synthetic crRNA with MS2 tracrRNA, SAM sgRNA plasmid, or SAM sgRNA lentiviral particles. When MS2 tracrRNA is combined with a single crRNA, it can be used to effectively induce short-term overexpression at levels comparable to, or better than, SAM sgRNA expression from commonly used plasmid and lentiviral delivery methods (Figure 3). A synthetic crRNA:MS2 tracrRNA system also offers some key benefits:
- Use in short-term assays or arrayed screening
- No cloning, sequencing, or in vitro transcription required
- Ability to combine individual crRNA into a pool for even more robust activation
CRISPRa comparison: synthetic crRNA with MS2 tracrRNA vs. expressed SAM system
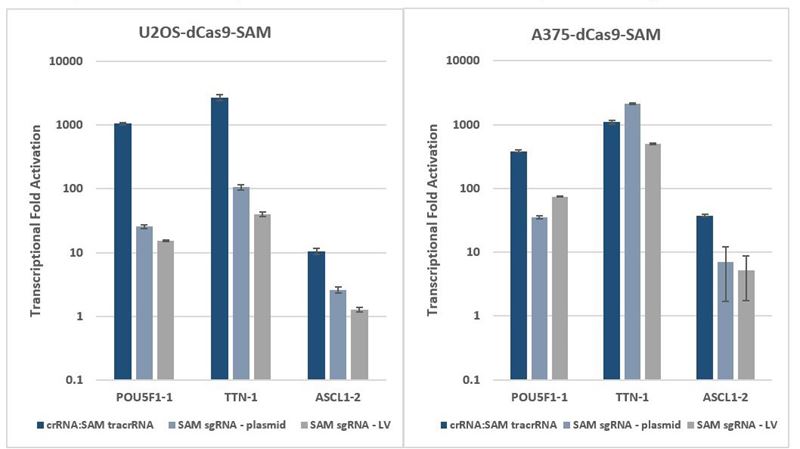
Figure 3. Activation with synthetic crRNA and MS2 tracrRNA: Cells were plated at 10,000 cells/well and were transfected using DharmaFECT 4 (U2OS-dCas9-SAM) or DharmaFECT 1 (A375-dCas9-SAM) Transfection Reagent with synthetic crRNA: MS2 tracrRNA (25 nM) targeting POU5F1, TTN, and ASCL1 genes. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR.
Activation with SAM sgRNA plasmid: Cells were plated at 10,000 cells/well and transfected with 200 ng of SAM sgRNA plasmids (containing two MS2 domains in the tracrRNA region1) targeting POU5F1, TTN, and ASCL1 genes using DharmaFECT kb Transfection Reagent. Cells were harvested 72 hours post-transfection (without antibiotic selection) and the relative gene expression was measured using RT-qPCR.
Activation with lentiviral sgRNA lentiviral particles: Cells were plated at 10,000 cells/well and were transduced with SAM sgRNA lentiviral particles (containing two MS2 domains in the tracrRNA region1) targeting POU5F1, TTN, and ASCL1 genes at a MOI of 0.3 to obtain cells with a single integrant. Cells were selected with 250 µg/mL zeocin for 48 hours prior to analysis with RT-qPCR.
The relative expression of each gene was calculated with the ∆∆Cq method using PPIB as the housekeeping gene and normalized to a non-targeting control.
Pooling multiple CRISPRa synthetic crRNA per gene further improves activation
When MS2 tracrRNA is complexed with an individual CRISPRa crRNA, robust activation is achieved. When complexed with multiple CRISPRa crRNAs targeting a single gene, enhanced activation levels can be achieved (Figure 4). Overall, pooling is a good strategy if maximal gene activation is desired, or if it is beneficial to decrease the scale of an experiment, for example, when performing analysis of multiple genes in an arrayed plate format.
Transcriptional activation with individual and pooled CRISPRa synthetic crRNA with MS2 tracrRNA
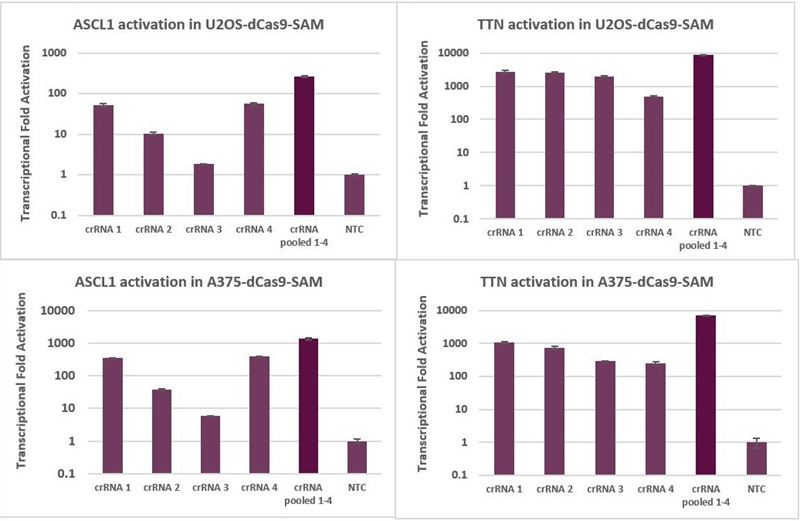
Figure 4. U2OS and A375 cells stably expressing dCas9-VP64 and MS2-p65-HSF1 were plated at 10,000 cells/well and transfected with either individual CRISPRa crRNA or pooled CRISPRa crRNAs, complexed with MS2 tracrRNA (25nM), using DharmaFECT 4 (U2OS-dCas9-SAM) or DharmaFECT 1 (A375-dCas9-SAM) Transfection Reagent. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using PPIB as the housekeeping gene and normalized to a non-targeting control.
References:
- S. Konermann et al., Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517, 583–588 (2015).
- M. A. Horlbeck et al., Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760 (2016).
Order Products
MS2 tracrRNA
Chemically synthesized MS2 tracrRNA for use with CRISPRa synthetic crRNA for fast and easy gene activation in stable cells containing the SAM transcriptional activation system.
CRISPRa synthetic crRNA
Predesigned synthetic guide RNA for over-expression of human and mouse genes using CRISPR activation. Just search for your gene! Available as pooled or individual reagents.
CRISPRa synthetic crRNA positive controls
Validated synthetic crRNA pool or individual for experimental optimization of transcription activation experiments.
CRISPRa synthetic crRNA non-targeting control
Non-targeting controls to evaluate baseline cellular responses to CRISPRa components in the absence of gene target-specific crRNA.
CRISPRa synthetic crRNA libraries
Arrayed collections of synthetic CRISPR RNA for overexpression screening of gene families, druggable, and whole human genome.
Cherry-pick libraries
Turn your gene list into a custom library of CRISPRa synthetic crRNAs.
Helpful Resources
Dharmacon™ Edit-R™ synthetic guide RNA resuspension protocol
This protocol is for the resuspension of Edit-R synthetic sgRNA, synthetic crRNA, synthetic tracrRNA, or synthetic SAM tracrRNA
CRISPRa: Transcriptional upregulation screening with genome-wide CRISPR activation
Building on the transformative impact of pooled screening using CRISPRCas9, researchers have developed catalytically inactive Cas9 (dCas9) and have demonstrated that it can be used to tune genetic output.
Gene overexpression: CRISPRa, cDNA or ORF?
Which gene expression technology is best for my experiment?
