Understanding tools that mimic or inhibit endogenous microRNA function
The endogenous RNAi pathway has been a primary research focus because of its role in post-transcriptional control of gene expression and its utility for siRNA and shRNA induced gene silencing. Undoubtedly, the function of endogenous microRNAs remains a subject of intensive research1. microRNAs can target hundreds of genes simultaneously2, induce subtle but reproducible shifts in gene expression and regulate of a variety of biological processes. Tools for elucidating microRNA function fall into two general categories: 1. reagents that mimic endogenous microRNA function and 2. reagents that inhibit endogenous microRNAs. As such, synthetic or expressed microRNA mimics and inhibitors are valuable tools for modulating microRNA expression, and can help to reveal the role of microRNAs in development, homeostasis and disease3-7.
Harnessing the endogenous microRNA pathway
A schematic of the endogenous microRNA pathway, with points of entry for the three categories of RNAi tools.
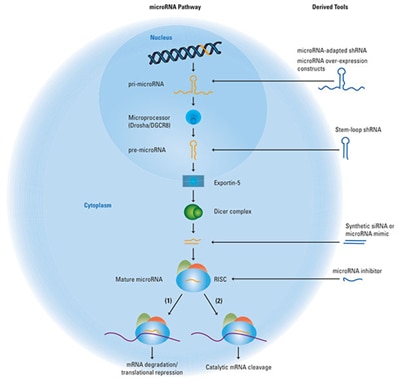
The endogenous microRNA pathway begins with genomic DNA transcription to produce pri-microRNA, which is then processed by the microprocessor complex containing the DROSHA protein and other proteins to a pre-microRNA hairpin structure and exported from the nucleus by Exportin-5. In the cytoplasm, the pre-microRNA is further processed by the Dicer complex to double-strand anti-parallel RNA. One strand of this RNA duplex will load into the RISC complex and act to target a mRNA transcript for down-regulation by either (1) seed-mediated mRNA translational repression or (2) catalytic mRNA cleavage mediated by high sequence complementarity. The blue molecules to the right of the image illustrate different classes of RNAi research tools and where they enter the endogenous RNAi pathway.
What are microRNA Mimics?
microRNA mimics are used experimentally to supplement endogenous microRNA expression, much like a cDNA can supplement gene expression, for gain-of-function studies. Generally, there are two types of microRNA mimetics: synthetic mimics and DNA-based over-expression constructs that produce short hairpin RNAs (shRNAs) based on microRNA sequences. Synthetic microRNA mimics are chemically synthesized, double-strand RNA molecules that are intended to mimic the transient duplexed product of Dicer complex processing. For synthetic mimics, like naturally occurring microRNAs, the two strands of a synthetic mimic separate, and the single-strand mature microRNA is incorporated into RISC to down-regulate mRNA transcripts. Synthetic mimics can be chemically modified to favor loading of the mature microRNA strand, and prevent loading of the passenger strand, as annotated in miRBase (www.mirbase.org). Lipid mediated transfection is the most common method of synthetic microRNA mimic delivery into cells.
For short or longer term expression, shRNAs based on microRNA sequences can be produced by DNA expression vectors. Depending on the vector, the shRNA-expressing DNA sequence may be extra-chromosomal or stably integrated into the genome, and expression can be either constitutive or inducible. Plasmid DNA expressing microRNA mimetics are transfected into the cells by standard methods.
What are microRNA Inhibitors?
When studying the functional role of a microRNA, both gain-of-function and loss-of-function observations should be made. The introduction of microRNA inhibitors into a biological system results in a loss-of-function assay with a predicted decrease in the function of the targeted endogenous microRNA. Two types of inhibitors are available: synthetic and expressed inhibitors. Synthetic inhibitors are generally a non-hydrolyzable, single-strand reverse complement to the mature microRNA. The mechanism of inhibition is likely mediated by irreversible binding of the inhibitor to mature microRNA-loaded RISC, thus preventing interaction of the mature microRNA to its endogenous mRNA targets. Expressed inhibitors (sometimes referred to as microRNA sponges) are generally artificial mRNA constructs with multiple microRNA sites that compete with natural mRNA target for binding of microRNAs2,8. Combination of these approaches and tools in loss-of-function assays increase the likelihood of observing the otherwise subtle phenotypes often associated with microRNA inhibition.
What Affects microRNA Function?
Unlike siRNA and shRNA tools, whose designs are generally not restricted by limitations of sequence space within the target mRNA, microRNA tools are based on sequences explicitly defined in miRBase (www.mirbase.org) and are not altered in order to preserve native function. Instead, alternative strategies are applied for enhancing activity of mimics and inhibitors. Both synthetic and expressed microRNA mimics function by increasing the abundance of mature microRNA sequences in a cell. To improve microRNA mimic function, chemical modifications and structural enhancement strategies can be applied to strongly bias for mature (or guide) strand RISC loading. Similarly, the function of microRNA inhibitors can be enhanced by modifying the inhibitor structure or using different chemical modifications to improve binding to mature microRNA9-11.
For expressed microRNAs, plasmid vectors can be engineered so that the active, mature microRNA sequences are expressed within the context of a universal microRNA scaffold. In this context, the native secondary structure of the microRNA scaffold promotes efficient, accurate Drosha and Dicer processing, and preferential loading of the active, mature strand of the microRNA with minimal activity from the opposite strand.
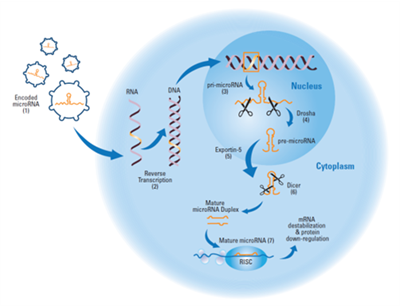
Borrowing the endogenous pathway
(1) The lentiviral particle binds to the host cell and delivers its engineered RNA genome, which includes encoded microRNA, to the cytoplasm. (2) The viral genome is reverse-transcribed in the cytoplasm (i.e. RNA to DNA). The DNA intermediate form is imported into the nucleus and is stably integrated into the host genome. (3) shMIMIC microRNA transgenes are transcribed in the same manner as endogenous microRNA genes. Native and over-expressed primary microRNAs are processed by the Drosha complex to yield the precursor microRNA (4), which is then shuttled out of the nucleus into the cytoplasm (5) and further processed by the Dicer complex into double-strand microRNA (6). One strand is selected for incorporation into the RNA-Induced Silencing Complex (RISC). microRNA-programmed RISC binds to target mRNA, causing transcript destabilization and subsequent down-regulation of protein expression (7).
Specificity of microRNA Targeting
The specificity of microRNAs is based on their mature sequence and is achieved through complementarity of target mRNAs with the seed region of the microRNA that is only six nucleotides in length. Consequently, one microRNA can target many different genes. The mammalian RNAi mechanism relies on this multiple targeting to regulate pathways, gene families, developmental checkpoints and observable phenotypes. Since microRNA mimics predominantly load and use the mature sequence, these tools mimic the specificity of the mature microRNA strand. In addition, some microRNAs are closely related in sequence (termed family members, (www.mirbase.org) and often have redundant function12. For that reason, there is potential for some cross-reactivity of inhibitors to several microRNAs when sequences are closely related13.
How are microRNA Mimetics and Inhibitors Delivered to Cells?
Synthetic microRNA mimics and inhibitors are typically delivered to cells by standard lipid-based reagents or electroporation. Ideally, the lipid reagent should be optimized for small RNA molecules. There are two options for the delivery DNA vectors expressing microRNA mimetics into cells. For plasmids, typical lipid transfection reagents or electroporation can be employed. Lentiviral particles are an excellent choice for difficult to transfect cells, and in situations where high efficiency is needed or multiple constructs per cell need to be delivered. Most modern vectors have selectable markers that allow the selective killing of any cells that were not successfully transfected or transduced, so that a pure culture can be developed.
Applications of microRNA Mimics and Inhibitors
microRNA mimics and inhibitors are essential tools to perturb microRNA function and assess the phenotypic consequences in gain- or loss-of-function assays. microRNA mimics and inhibitors can be used in conjunction with microRNA and gene expression profiling for identification of specific microRNA-gene target relationships. In addition, several screens have been performed that demonstrate microRNA involvement in biological systems such as cancer and viral infection6,7. While both microRNA mimics and inhibitors have potential for studies in vivo, microRNA inhibitors in particular have shown promise in therapeutic development14-17.
References
- Huntzinger, E. and Izaurralde, E. (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Reviews Genetics 12(2):99-110.
- Friedman, R.C. et al. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Research 19(1):92-105.
- Lim, L. et al. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769-773.
- Krutzfeldt, J. et al. (2005) Silencing of microRNAs in vivo with 'antagomirs'. Nature 438:685-689.
- Horwich, M.D. and Zamore, P.D. (2008) Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nature Protocols 3:1537-1549.
- Wang, Y. et al. (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nature Genetics 40(12):1478-1483.
- Santhakumar, D. et al. (2010) Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. PNAS 107(31):13830-13835.
- Thomas, M. et al. (2010) Desperately seeking microRNA targets. Nature Structural and Molecular Biology 17(10):1169-1174.
- Vermeulen, A. et al. (2007) Double-strand regions are essential design components of potent inhibitors of RISC function. RNA 13(5):723-730.
- Davis, S. et al. (2006) Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Research 34:2294-2304.
- Ørom, U.A. et al. (2006) LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 372:137-141.
- Gregory, P.A. et al. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology 10:593-601.
- Robertson, B. et al. (2010) Specificity and functionality of microRNA inhibitors. Silence. 1(1):10.
- Montgomery, R. and van Rooij, E. (2011) Therapeutic advances in MicroRNA targeting. Journal of Cardiovascular Pharmacology. 57(1):1-7.
- Elmen, J. et al. (2008) Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Research 36(4):1153-1162.
- Trang, P. et al. (2011) Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Molecular Therapy 19(6):1116-1122.
- Petri, A. et al. (2009) MicroRNA Silencing in primates: towards development of novel therapeutics. Cancer Research 69(2):393-395.
Order Products
microRNA Product Overview
Up-regulate or suppress endogenous mature microRNA function with rationally designed synthetic and expressed microRNA modulation reagents.
miRIDIAN microRNA mimics
Supplement endogenous microRNA activity to study gain-of-function effects.
Custom microRNA Modulation Tools
Modify an existing microRNA mimic or inhibitor, target a novel microRNA, or request an Accell microRNA Inhibitor or Stabilized Mimic for special applications.
miRIDIAN microRNA Hairpin Inhibitors
Innovative microRNA inhibitor design suppresses microRNA activity to study loss-of-function effects and allows for superior potency and longevity. SMARTchoice Platform Maximize expression with your choice of constitutive and inducible promoters!
Helpful Resources
Screening Selection Guide
Short-term knockdown, stable/long-term knockdown, modulating of non-coding RNA and gene expression selection guidelines.
Dharmacon microRNA Selection Guide
A quick tool to help determine the ideal Dharmacon microRNA product line and format for your needs.
