Controlling the timing of the editing events
A necessary component of any gene editing experiment is the Cas9 nuclease. This enzyme is responsible for generating the cut in the dsDNA that allows either non-homologous end joining (NHEJ) or homology-directed repair (HDR) to occur and results in the change in the gene DNA sequence. However, controlling the timing of the experiment can be a critical part of the overall success or failure of a gene editing experiment. This can be especially true when trying to set up a pooled screen, generate a stable cell line without risk of increased metabolic load, or in cases where it is desirable to integrate the Cas9 in a precursor state and perform an experiment in a later derived cell state.
An inducible Cas9 expression vector can provide experimental flexibility
Dharmacon Edit-R Inducible Lentiviral Cas9 Nuclease provides the temporal control necessary to ensure editing only occurs when it is required. Utilizing the tight, highly responsive Tet-On® 3G doxycycline inducible promoter and built on the platform of our existing human codon optimized constitutive Cas9 nuclease vectors, this vector provides robust expression of the Cas9 nuclease when induced and minimal leak in the off state. Couple this control with the precise editing of Edit-R lentiviral and synthetic guide RNAs to result in an unparalleled level of options for your next CRISPR-Cas9 gene editing experiment. Available as either ready-to-use purified high-titer lentiviral particles or a purified plasmid format ready for packaging in the lab, this new vector will give you the precision you require.
Dose response for doxycycline in inducible U2OS-Cas9 cells
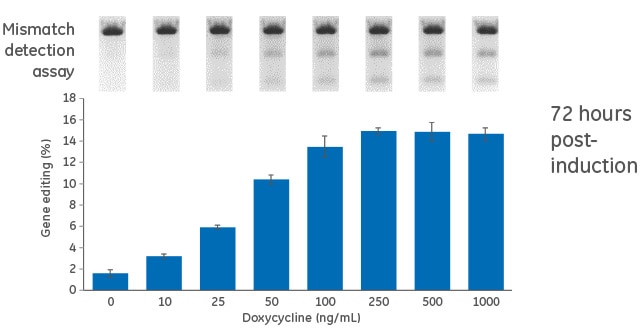
Figure 1. Cells were transduced with the inducible (TRE3G-Cas9) Cas9 expression lentiviral particles at an MOI of 0.3, and selected with 10 µg/mL blasticidin in tetracycline-free medium for 10 days. Cas9-stable cells were then transduced with PPIB-sgRNA lentiviral particles at an MOI of 0.3. Cells were selected with 2 µg/mL puromycin tetracycline-free medium for 4 days, suspended with trypsin and seeded in a 96-well plate in medium containing increasing concentrations of doxycycline (0 to 1000 ng/mL). The cells were incubated for 72 hours, lysed and analyzed for indel formation using a DNA mismatch detection assay with T7EI. Upper panel, representative gel image of the DNA mismatch detection assay with T7EI for PPIB targeted amplicon; lower panel, mean ± standard deviation of the estimated percentage of gene editing from three independently treated wells.
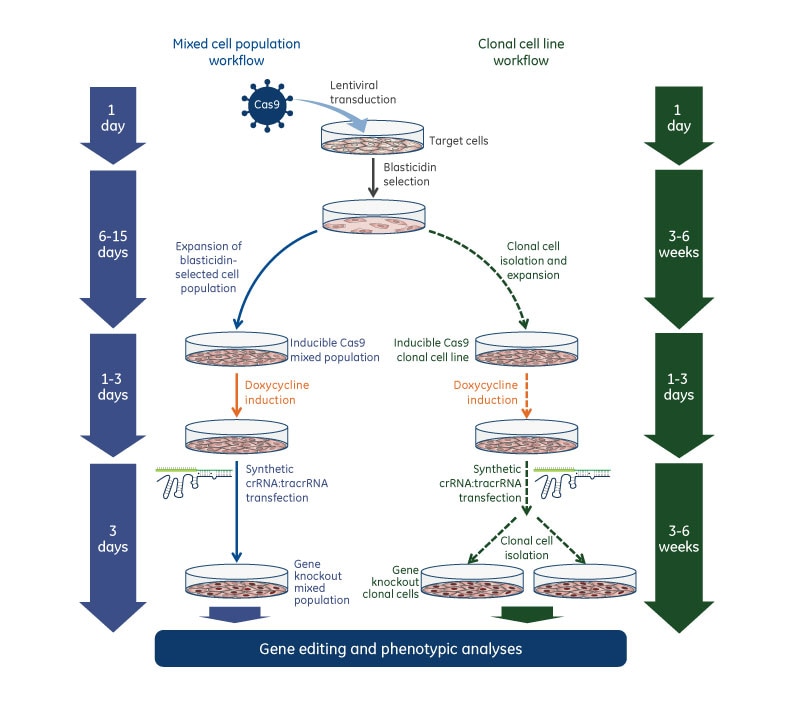
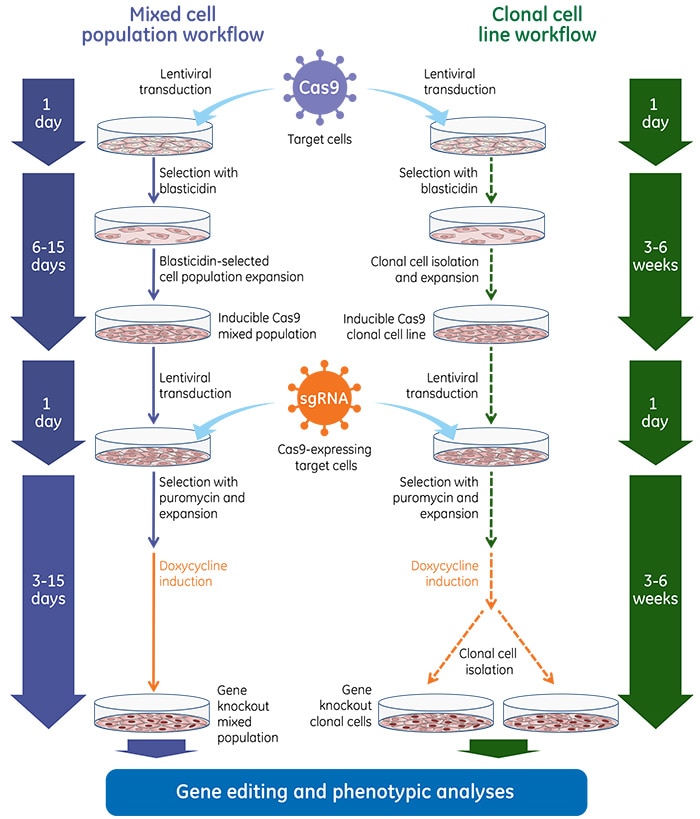
Gene knockout workflows with Edit-R Inducible Lentiviral Cas9 Nuclease
Once inducible Cas9 cell lines are generated, these cells are transduced with gene-specific sgRNA lentiviral particles and subsequent gene knockouts can be obtained by treatment with doxycycline at a predefined concentration. Below are general experimental workflows to generate stable cell lines carrying the inducible Cas9 followed by transduction with sgRNA lentiviral particles and induction with doxycycline for phenotypic analysis of gene knockout.
Gene knockout workflow with Edit-R Inducible Lentiviral Cas9 Nuclease and Edit-R synthetic crRNA:tracrRNA
Gene knockout workflow using the Edit-R Inducible Lentiviral Cas9 Nuclease with Edit-R Lentiviral sgRNA
Order Products
Edit-R Lentiviral Cas9 Nuclease Reagents
Lentiviral CRISPR-Cas9 components for robust gene editing in biologically relevant cell types
Cas9 Nucleases
Configurable expression constructs or DNA-free options for optimal Cas9 expression
Helpful Resources
View tech manual
Edit-R Inducible Lentiviral Cas9 Technical Manual
